Translate this page into:
Molecular characterization and epidemiology of methicillin-resistant Staphylococcus aureus isolated from clinical samples in Sokoto, Nigeria

*Corresponding author: Asiya Imam Umar, Department of Medical Microbiology, Usmanu Danfodiyo University, Sokoto, Nigeria. asiya.imam@udusok.edu.ng
-
Received: ,
Accepted: ,
How to cite this article: Umar AI, Manga SB, Baki AS, Uba A. Molecular characterization and epidemiology of methicillin-resistant Staphylococcus aureus isolated from clinical samples in Sokoto, Nigeria. Adesh Univ J Med Sci Res 2023;5:17-24.
Abstract
Objectives:
Methicillin-resistant Staphylococcus aureus (MRSA) is a major public health threat and a cause of hospital-acquired and community-acquired infections. This study was undertaken to investigate antimicrobial resistance pattern, the genetic lineage, presence of S. aureus protein A (SPA) gene, and staphylococcal chromosomal cassette mec (SCC mec) types of MRSA isolated from clinical samples sent for microbiological test in major government hospitals in Sokoto.
Material and Methods:
A total of 90 S. aureus MRSA isolates were collected and confirmed using standard microbiological techniques. Antibiotic susceptibility testing was done using the disk diffusion method; mecA detection and sequencing were carried out. Phylogenetic reconstruction was also done using the molecular evolutionary genetics analysis X software and phylogeny tree constructed by Neighbor-Joining method. SCC mec typing and SPA detection were also done.
Results:
Of the 90 S. aureus isolates, 42 were found to be MRSA using the cefoxitin disk diffusion, the most potent antibiotic against them was quinupristin/dalfopristin with 83.3% followed by rifampicin with 81.0% and 6 clindamycin with a 71.4%. With 78.6% of the isolates showing resistance to the fluoroquinolone antibiotic ciprofloxacin, tetracycline and gentamicin came in second and third, with 64.3% and 61.9% of isolates showing resistance, respectively. Most of the MRSA isolates were resistant to more than three antibiotics. Polymerase chain reaction showed 36 (85.7%) harbored the mecA gene and of the 36 mecA positive isolates, phylogenetic reconstruction of representative MRSA sequences showed that MRSA sequences in this study clustered in two closest clades suggesting a possible horizontal transfer. Of the 36 isolates, 23 were SCC mec type I, ten were type IV, and three were non-typeable, while the SPA gene was detected in all the isolates amplified.
Conclusion:
The use of phenotypic and molecular methods in this study provided useful information on antibiotic resistance profile, epidemiology, and molecular characteristics of MRSA isolates in Sokoto Nigeria. The information provided could help in monitoring the evolution of MRSA strains in Nigeria over time.
Keywords
Methicillin-resistant Staphylococcus aureus
mecA
Staphylococcus aureus protein A
Staphylococcal chromosomal cassette mec
Sequences
INTRODUCTION
Hospital-and community-acquired infections now pose a serious threat due to methicillin-resistant Staphylococcus aureus (MRSA) illnesses.[1,2] Endocarditis, osteomyelitis, necrotizing pneumonia, bacteremia, septicemia, meningitis, food poisoning, and toxic shock syndrome are only a few of the ailments that the organism has been linked to.[3] More than 90% of Staphylococcus strains are resistant to penicillin as a result of a change in the penicillin-binding protein 2a (PBP2a), which has a decreased affinity for b-lactam antibiotics,[4] followed by increasing resistance to methicillin, aminoglycosides, macrolides, and lincosamide.[5] The mecA gene on a staphylococcal chromosomal cassette mec (SCC mec) encodes the PBP2a protein. MRSA epidemiologic typing and molecular characterization are critical for tracking the emergence and spread of new epidemic clones and determining intervention strategies.[6] Hospitals are no longer immune to the threat of MRSA colonization and infection. Different typing techniques, such as multilocus sequence typing, SCC mec typing, and S. aureus protein A (SPA) detection and typing, can be used to identify MRSA lineages and strains. The data gathered in this way can be epidemiologically helpful for tracking outbreaks, determining the most likely source of colonization (such as connected with livestock or humans), and differentiating between community and hospital strains: MRSA was formerly only linked with medical facilities, but it has since become a significant contributor to community-related infections and has established reservoirs in both of these environments. This shows that the global spread of community-associated MRSA (CA-MRSA) strains has altered the epidemiology of MRSA. Hospital-associated MRSA (HA-MRSA) differs genetically from CA-MRSA in that the latter is less resistant to non-lactam antibiotics, has a smaller form of SCC mec, and frequently produces the cytotoxin Panton-Valentine leukocidin.[7] Determining the SCC mec type is critical in determining whether the clone was obtained in a hospital or in the community. The SCC mec types: I, II, and III are associated with HA-MRSA strains while types IV and V are considered as community-associated MRSA strains.[7] SPA typing is also frequently used and based on typing of protein A. The SPA typing method is based on DNA sequencing of short-sequence repeats in the staphylococcal protein A gene’s polymorphic conserved X region.[8] This study aimed to investigate the genetic diversity and epidemiology of MRSA isolates identifying the prominent clones circulating in health-care settings in Sokoto, Nigeria.
MATERIAL AND METHODS
Bacterial isolates and data collection
In this cross-sectional analytical study, a total of 90 S. aureus isolates were collected from different sources, including wound swab, nasal swabs, urine, pus, urethral swabs, high vaginal swabs (HVSs), endocervical swabs (ECSs), ear swabs, catheter tips, and wound aspirate and transported to the research laboratory immediately.[9] Furthermore, information concerning the isolates, that is, sample type, age (0–60 years), sex (male or female), and in- or outpatient and clinical diagnosis was collected.
Bacterial culture and identification of MRSA strains
MRSA isolates were identified phenotypically by colony morphology on blood agar and Mannitol salt agar base, Gram stain, catalase, coagulase, and DNAse tests. The methicillin-resistant strains were identified by the disk agar diffusion method using a cefoxitin disk/30 μg.
Antibiotic susceptibility testing
Standard inoculum was prepared by making a direct saline suspension of isolated colonies selected from an 18-h agar plate incubated at 37°C. The suspension was adjusted to achieve a turbidity equivalent to a 0.5 McFarland (1–2 × 108 colony-forming unit/mL). It was then observed, using adequate light to visually compare the inoculum tube and the 0.5 McFarland standards against a card with a white background and contrasting black lines. Antibiogram was done in accordance to Clinical and Laboratory Standard Institute. Commercially prepared antibiotic disks, quinupristin/dalfopristin (15 μg), clindamycin (2 μg), gentamicin (30 μg), ciprofloxacin (5 μg), rifampicin (5 μg), teicoplanin (2 μg), tetracycline (30 μg), and erythromycin (15 μg) were placed on the inoculated Mueller–Hinton agar 25 mm away from each other. The plate was then incubated at 35°C for 18–24 h after which the zones were read using the interpretation chart provide d by the clinical laboratory standard institute.[10] In this study, multidrug resistance is considered to be resistance to at least three antibiotics of different classes.
DNA extraction and quantification
Genomic DNA was extracted from overnight fresh pure cultures on blood agar using Qiagen (USA) DNA extraction kit following Manufacturer’s protocol.
Identification of MRSA strains
The MRSA strains were confirmed by the detection of mecA gene using polymerase chain reaction (PCR) as described by Murakami et al.[11] The mecA product purification was done using DNA clean and concentrator kit (Zymo Research).
Sequencing and phylogenetic analysis of mecA products
The mecA purified products were sequenced using Sanger sequencing at Inqaba Biotech South Africa. The sequences received were accessed using molecular evolutionary genetics analysis X software (MEGA X); thereafter, the FASTA format of the sequences was copied and pasted on the national center for biotechnology information basic local alignment search tool for nucleotides database command option to determine the similarity between the sequences obtained and sequences deposited in the Genbank.[8] The product of BLAST sequences was aligned by ClustalW pairwise and multiple alignment s using MEGAX software. Best DNA/ protein model for the reconstruction of phylogenetic tree was determined and then the phylogeny tree constructed by Neighbor-Joining method tree.
PCR identification of SCC mec types
SCC mec types (I-V) were identified by multiplex PCR as described by Boye et al.[12] The primers used in this multiplex PCR assay are shown in [Table 1].
| # | Gene | Oligo Name | Primer sequence 5'-3' Reference | Length (bp) | Reference |
|---|---|---|---|---|---|
| 1 | mecA | mecA1F | AAAATCGATGGTAAAGGTTGGC | 533 | (Murakami et al., 1991) |
| mecA2R | AGTTCTGCAGTACCGGATTTGC | ||||
| 2 | SCC mec multiplex | b/ccrA2F-B | ATTGCCTTGATAATAGCCYTCT | 937 | (Ito et al., 2001) |
| a3/ccrA2R-B | TAAAGGCATCAATGCACAAACACT | ||||
| ccrCF/ccrC | CGTCTATTACAAGATGTTAAGGATAAT | 518 | (Ito et al., 2001) | ||
| ccrCR/ccrC | CCTTTATAGACTGGATTATTCAAAATA | ||||
| 1272F1/IS1272 | GCCACTCATAACATATGGAA | 415 | (Boye et al., 2007) | ||
| 1272R1/IS1272 | CATCCGAGTGAAACCCAAA | ||||
| 5RmecAF/IS431 | TATACCAAACCCGACAACTAC | 359 | (Hadyeh et al., 2019) | ||
| 5R431R/IS431 | CGGCTACAGTGATAACATCC | ||||
| VF | GAACATTGTTACTTAAATGAGCG | 325 | (Fri et al., 2020) | ||
| VR | TGAAAGTTGTACCCTTGACACC | ||||
| 3 | SPA | 1095F/SPA-F | AGACGATCCTTCGGTGAGC | 200–400 | (Hadyeh et al., 2019) |
| 1017R/SPA-R | GCTTTTGCAATGTCATTTACTG |
SCC mec: Staphylococcal chromosomal cassette mec, SPA: Staphylococcus aureus protein A
Detection of SPA gene
All MRSA isolates were subjected to PCR to detect the presence of the SPA gene using the PCR primers and cycling as previously described by Hadyeh et al. [Tables 1 and 2].[8]
| # | Target Gene | PCR program (Temp°C/Time)-35 cycles | Reference | ||||
|---|---|---|---|---|---|---|---|
| Initial denaturation | Denaturation | Annealing | Extension | Final extension | |||
| 1 | mecA | 95°C/5 min | 95°C/30 s | 58°C/30 s | 72°C/80 s | 72°C/10 min | (Murakami et al., 1991) |
| 2 | SCC mec | 95°C/5 min | 94°C/30 s | 55°C/30 s | 72°C/80 s | 72°C/10 min | (Boye et al., 2007) |
| 3 | SPA | 95°C/5 min | 95°C/30 s | 58°C/30 s | 72°C/45 s | 72°C/10 min | (Hadyeh et al., 2019) |
SCC mec: Staphylococcal chromosomal cassette mec, SPA:Staphylococcus aureus protein A
PCR assay and visualization
All PCR reactions were optimized and carried out by the Basic Gradient Thermocycler using Mastermix (BioLabs New England) according to the manufacturer’s instructions, [Table 2]. Amplicons for all the characterized genes were analyzed electrophoretically in 1.5% agarose gels and visualized by UV light using UV visible doc imager (Biorad imager, Germany).
Methods and indices for data analysis
Data obtained were presented using tables, percentages, and Statistical Package for the Social Sciences Windows version 23. The degree of confidence was set at 95% (P ≤ 0.05).
RESULTS
Bacterial isolates and study population
Based on the prevalence of MRSA among the isolates collected from the two hospitals, Specialists Hospital accounted for 20 (57.1%) using cefoxitin disk diffusion, while Usmanu Danfodiyo University Teaching Hospital (UDUTH) had 22 (36.7%). The highest prevalence was observed in the age group of 21–30 years where 13 (56.5%) of the 23 isolates were found to be MRSA, this is followed by 1–10 years age group 8 (47.1%) 17 isolates being MRSA. The age group with the least prevalence is 41–50 years age group only 1 (14.3%) out of seven isolates being MRSA. Nineteen (39.6%) were isolated from the male gender and 23 (54.8%) from the females. Of the 42 patients whose samples were used in this research, 28 (51.9%) were on admission while the remaining 14 (34.1%) were out patients. Based on sample type, 100% prevalence seen in nasal swab, urethral swab, and pus, followed by urine with 53.8%, HVS 42.1%, ECS 25%, wound swab 46.7%, wound aspirate 25%, and catheter tip yielded a prevalence of 20%, while no resistant isolate was recovered from ear swab.
Antimicrobial susceptibility patterns
The most potent antibiotic was quinupristin/dalfopristin with 83.3% of all the tested isolates being sensitive and an inhibition zone of ≥19 mm, it is followed by rifampicin with 81.0% inhibition zone of ≥20 mm and clindamycin with a 71.4% sensitivity rate and inhibition zone of ≥21 mm. The least activity was shown to be in fluoroquinolone antibiotic ciprofloxacin for which 78.6% of the isolates demonstrated phenotypic resistance with an inhibition zone of ≤15 mm, it is followed by erythromycin with inhibition zone of ≤13 mm (64.3%), tetracycline with inhibition zone of ≤14 mm (61.9%), and gentamycin with inhibition zone of ≤12 mm (61.9%); this is shown in [Figure 1].
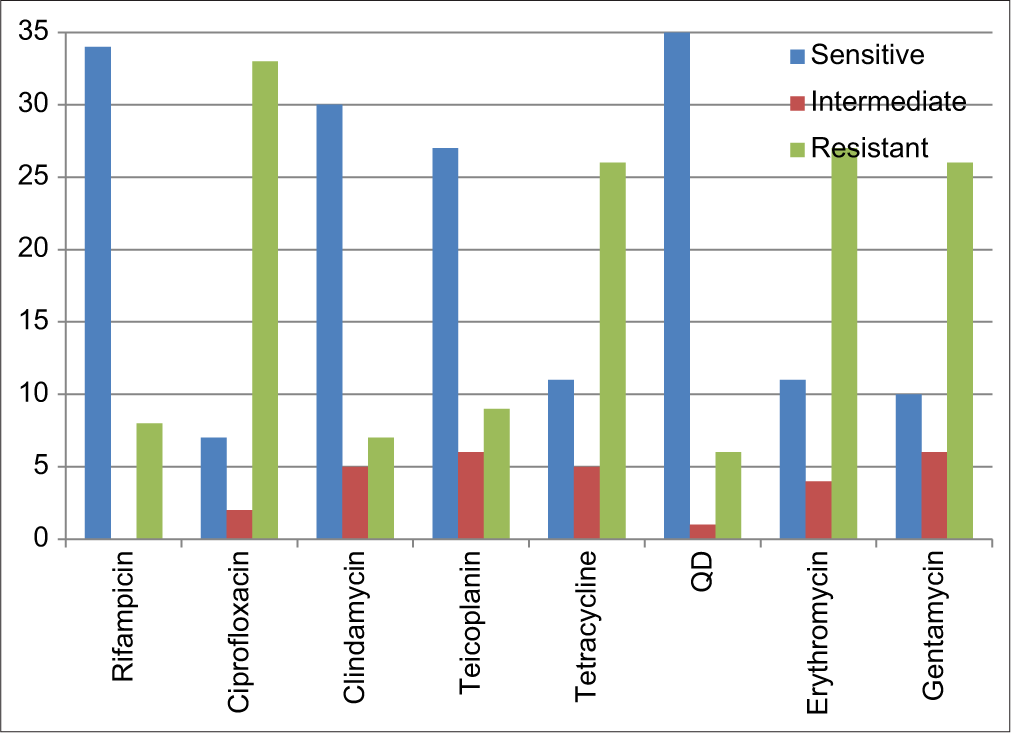
- Antimicrobial profile of the Methicillin-resistant Staphylococcus aureus isolates to different antibiotics.
Confirmation of methicillin resistance by detection of the mecA gene
Of the 42 isolates found to be methicillin-resistant phenotypically using cefoxitin disk diffusion, only 36 were confirmed to be genotypically methicillin resistant by the detection of the mecA gene using PCR targeting the mecA gene yielding 533 bp product.
Phylogenetic analysis of mecA products
[Table 3] depicts the results for the mecA nucleotide sequences obtained, with all five samples showing best hit with S. aureus strain UNC_SaCF11 accession number CP089162.1, 100% query cover, and sequence identity of 97.75–100%. The evolutionary relationships of taxa to our samples were determined and the evolutionary history was inferred using the Neighbor-Joining method and optimal tree is shown for each sample. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. This analysis involved 39 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Non-coding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 101 positions in the final dataset. Evolutionary analyses were conducted in MEGA X, as shown in [Figure 2].
| Sample ID | Blastn results for the mecA nucleotide sequences | Query cover % | Sequence identity % | Accession number |
|---|---|---|---|---|
| Best hit | ||||
| mecA 1 | S. aureus strain UNC_SaCF11 | 100 | 98.73 | CP089162.1 |
| mecA 3 | S. aureus strain UNC_SaCF11 | 100 | 100 | CP089162.1 |
| mecA 8 | S. aureus strain UNC_SaCF11 | 100 | 97.75 | CP089162.1 |
| mecA 9 | S. aureus strain UNC_SaCF11 | 100 | 98.73 | CP089162.1 |
| mecA 10 | S. aureus strain UNC_SaCF11 | 100 | 98.73 | CP089162.1 |
Blastn: Basic local alignment search tool for nucleotides. S. aureus: Staphylococcus aureus

- Evolutionary relationship of Taxa using the Neighbor-Joining method.
The evolutionary history was inferred using the Neighbor-Joining method and optimal tree is shown in [Figure 3]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA X.

- Evolutionary relationships of our clustered sequences.
Molecular characterization and typing of isolates
SCCmec typing
Among all the 36 mecA positive MRSA isolates, 23 isolates belonged to SCC mec type I (63.9%) and ten belonged to SCC mec type IV (27.8%). The isolates were identified as CAMRSA when carrying the SCC mec type IV and HA-MRSA when carrying the SCC mec type I. Types II, III, and V were not amplified (implying their absence in the isolates), while three of the isolates were non-typeable (NP) [Table 4].
| Center | SHS | UDUTH | Total |
|---|---|---|---|
| SCC mec type | |||
| I | 13 | 10 | 36 |
| IV | 2 | 8 | |
| NT | 3 | 0 | |
| SPA | |||
| Positive | 18 | 18 | 36 |
| Negative | 0 | 0 |
SCC mec: Staphylococcal chromosomal cassette mec, SPA:Staphylococcus aureus protein A, MRSA: Methicillin-resistant Staphylococcus aureus, SHS: Specialists Hospital, UDUTH: Usmanu Danfodiyo University Teaching Hospital
SPA gene detection
The SPA gene was detected in all the 36 mecA positive MRSA isolates tested.
DISCUSSION
Oxacillin or methicillin-resistant (MRSA) isolates are among the major pathogens causing infections in the world, leading to the emergence of and disseminating increasingly virulent and multiresistant strains, causing both nosocomial and community-acquired infections. Rapid and accurate detection of methicillin resistance in S. aureus is important for the use of appropriate antimicrobial therapy and for the control of nosocomial spread of MRSA strains.[13] This study showed that 42 (44.2%) of the isolates obtained were methicillin resistant by the cefoxitin disk diffusion method. This finding is higher than findings of Adetayo et al.,[14] Abdullahi and Iregbu,[15] and Breves et al., who reported a prevalence of 30.4% in Ibadan, 26.9% in Abuja, and 31.4% in Brazil, respectively. The rate of 44.2% from this study is however similar to what was reported by Adeiza et al.,[16] in Sokoto with 46.9%, Ariom et al.,[17] in Abakaliki with 43.4%, and Samson and Anthony[18] in Benin City with 79%, indicating that MRSA is ever increasing. This is further corroborated by findings of Abubakar and Sulaiman,[19] who, in a systematic review of MRSA infections in Nigeria, reported an increase from 18.3% (2009) to 42.3% (2013). It is clear that MRSA has become a global nosocomial pathogen with attendant therapeutic problems and warrant urgent infection awareness, considering the common practice of unregulated sale of antimicrobial agents and movement of people which may result in rapid dissemination.
The antibiogram pattern of the MRSA isolates showed that 39 (92.9%) were multi drug resistant and only 3 (7.1%) were to 2 or less antibiotics. The pattern showed a 78.6%, 64.3%, and 61.9% resistance to ciprofloxacin, erythromycin, and tetracycline, respectively, while the most potent of the antibiotics tested were quinupristin/dalfopristin with 83.3%, rifampin with 81.0%, and clindamycin with 71.4%. However, Mofolorunsho et al.,[20] in Anyingba, reported 54% resistance to erythromycin and augmentin, and sensitivity to gentamicin, ofloxacin, and ciprofloxacin as 100%, 81.8%, and 72.7%, respectively, while Ariom et al.,[17] in Abakaliki, reported that the clinical isolates were completely resistant (100%) to ceftazidime, tetracycline, and penicillin and that gentamicin and ciprofloxacin were the most effective antibiotics. Another research conducted by Imam et al.,[21] in Sokoto showed a 100% resistance to ceftazidime, cloxacillin, and augmentin, while the most potent of the antibiotics tested were nitrofurantoin, quinupristin/dalfostrin, and chloramphenicol with 96.7%, 95.7%, and 86%, respectively. Bunza et al.,[22] also in Sokoto, reported that 40% of the S. aureus isolates were susceptible to clindamycin, 64% to ciprofloxacin, 57% to erythromycin, 71% to gentamicin, 34% to cefoxitin, 46% to quinupristin/ dalfostrin, and 58% to tetracycline and sulfamethoxazole. The pattern of resistance shown by S. aureus to many groups of antimicrobial agents in this research represents a serious concern in therapeutic option available to the clinician in managing such infections and, further, confirms various literatures that S. aureus is a multidrug resistant bacteria. However, the potency of quinopristin/dalfostrin, rifampin and clindamycin seen in this study is an indication that physicians can still prescribe this antibiotic based on empirical therapy when needed, especially for urgent infections.
Phylogenetic reconstruction of mecA gene in combination with PCR detection of mecA is important in defining and understanding the molecular epidemiology of MRSA strains circulating in an area by comparing them with existing strains deposited in Gene bank.[23] Nucleotide sequence analyses of the representative mecA positive samples have indicated high percentages (97.75–100) of sequence identity with S. aureus strain UNC_SaCF accession number CP089162.1. The strain was isolated from a clinical sputum sample of a patient in Boston USA, and all our isolates were from clinical samples. On reconstruction of a phylogenetic tree, it was discovered that four of, our MRSA strains clustered on the same clade and the nucleotide sequences analyses of several mecA genes from different species used in this study have revealed that the mecA gene is much more conserved among staphylococcal species of human origin. The phylogenetic reconstruction in this study showed that all the mecA positive MRSA strains clustered into two closest clades, is suggestive of a possible transmission between them but the exact direction of the transmission could not be ascertained from this study (cross-sectional study). This result also suggests that there might be a possible horizontal mecA gene transfer between the people and also agrees with the general assumption that infectious disease spread can be influence by both local and international travel. In agreement to this, Zhou et al.[24] reported that increasing local and international travel facilitates the transmission of various multidrug-resistant bacteria including MRSA across continents.
Among the 36 mecA positive isolates, 23 (63.9%) were SCC mec type I, 10 (27.8%) SCC mec type IV, type II, III, and V were not detected, while 3 (8.3%) isolates were negative and designated NP. The SCC mec typing of the MRSA strains in this study detected only SCC mec types I and IV. SCC mec types I and II have historically been associated with multiresistance, that is, resistance to more than three antimicrobials[25] and that majority of the HA-MRSA carries SCC mec type I, II, or III.[26] The findings in this study are similar to what was reported within the country: Ibadan,[27] Ogun,[25] and Edo[28] and outside; Iran,[29] Zambia,[30] and India.[31] SCC mec type IV was the second most dominant type seen in this study and is usually associated with CA-MRSA. However, it has been increasingly found in HA-MRSA. Moreover, the HA-MRSA strains with SCC mec type IV are multidrug resistant while CA-MRSA strains are generally more susceptible.[30] Others however have attributed their increasing occurrence as due to their small size that can spread among S. aureus isolates collected from hospitals and communities.[32] Some studies, however, have reported other SCC mec types as predominating in their respective regions.[33] This can be attributed to the fact that Staphylococcal genomes seem to change continuously as genetic elements move in and out, even though no mechanism of transfer has been found responsible for moving SCC elements between different staphylococcal species yet. The presence of SCC mec type IV in considerable percentages of S. aureus isolated from MRSA cases from clinical isolates is a cause for concern. The sources of CA-MRSA in the hospital environment could be the patients, MRSA carrier individuals attending the hospital or the medical staff.
Protein A (SPA) is one of the virulence factors on the surface of S. aureus, that prevents the phagocytosis of the bacteria by the immune system.[34] In this study, the presence of the protein was detected in all the 36 mecA positive isolates.
CONCLUSION
The use of phenotypic and molecular methods in this study provided useful information on antibiotic resistance profile, epidemiology, and molecular characteristics of MRSA isolates in Sokoto Nigeria. The information provided could help in monitoring the evolution of MRSA strains in Nigeria over time.
Recommendations
For adequate information and proper treatment of S. aureus infections, it is of paramount importance to conduct further studies to help understand the trends in the antibiotic-resistance patterns of MRSA and other pathogenic bacteria in the study area. Knowledge about the nature and number of MRSA clones that are disseminating is required to implement any strategies to control the transmission of MRSA within hospitals.
Declaration of patient consent
Patient’s consent not required as patients identity is not disclosed or compromised.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Clinical and economic impact of methicillin resistance in patients with Staphylococcus aureus bacteremia. Diagn Microbiol Infect Dis. 2005;52:113-22.
- [CrossRef] [PubMed] [Google Scholar]
- Methicillin-resistant Staphylococcus aureus on orthopaedic wards: Incidence, spread, mortality, cost and control. J Bone Joint Surg Br. 2006;88:812-7.
- [CrossRef] [PubMed] [Google Scholar]
- Methicillin-resistant Staphylococcus aureus (MRSA): One health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255-65.
- [CrossRef] [PubMed] [Google Scholar]
- The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178-82.
- [CrossRef] [PubMed] [Google Scholar]
- Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: Report of a clinical failure. Antimicrob Agents Chemother. 2005;49:1222-4.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiological and molecular characterization of community and hospital acquired Staphylococcus aureus strains prevailing in Shenyang, Northeastern China. Braz J Infect Dis. 2013;17:682-90.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of community-and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976-84.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of methicillin resistant Staphylococcus aureus in West Bank-Palestine. Front Public Health. 2019;7:130.
- [CrossRef] [PubMed] [Google Scholar]
- Isolation and identification of vancomycin resistant Staphylococcus aureus from post operative pus sample. Al Ameen J Med Sci. 2011;4:152-68.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement, M100-S30. Vol 4. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- [Google Scholar]
- Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240-4.
- [CrossRef] [PubMed] [Google Scholar]
- A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec Types I-V. Clin Microbiol Infect. 2007;13:725-7.
- [CrossRef] [PubMed] [Google Scholar]
- Methicillin-and vancomycin-resistant Staphylococcus aureus in health care workers and medical devices. J Brasil Patol Med Lab. 2015;51:143-52.
- [CrossRef] [Google Scholar]
- Prevalence of methicillin resistant Staphylococcus aureus from Clinical Specimens in Ibadan, Nigeria. Int J Eng Sci. 2014;3:2319.
- [Google Scholar]
- Methicillin-resistant Staphylococcus aureus in a central Nigeria tertiary hospital. Ann Trop Pathol. 2018;9:6-10.
- [CrossRef] [Google Scholar]
- Prevalence, risk-factors, and antimicrobial susceptibility profile of methicillin-resistant Staphylococcus aureus (MRSA) obtained from nares of patients and staff of Sokoto state-owned hospitals in Nigeria. GMS Hyg Infect Control. 2020;15:Doc25.
- [CrossRef] [Google Scholar]
- Detection and phenotypic characterization of methicillin-resistant Staphylococcus aureus from clinical and community samples in Abakaliki, Ebonyi State, Nigeria. Afr Health Sci. 2019;19:2026-35.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of multi-drug resistant Staphylococcus aureus in clinical specimens obtained from patients attending the University of Benin Teaching Hospital, Benin City, Nigeria. J Nat Sci Res. 2013;3:154-9.
- [Google Scholar]
- Prevalence, trend and antimicrobial susceptibility of Methicillin Resistant Staphylococcus aureus in Nigeria: A systematic review. J Infect Public Health. 2018;11:763-70.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus prevalence and antibiotic susceptibility profile in anyigba, north-central Nigeria. Am J Infect Dis. 2015;11:93-7.
- [CrossRef] [Google Scholar]
- Prevalence and antibiogram of Staphylococcus aureus isolated from clinical samples in Sokoto metropolis. Int J Innov Res Dev. 2018;7:1-7.
- [Google Scholar]
- Antibiotic susceptibility pattern of Staphylococcus aureus isolated from clinical samples in specialist hospital, Sokoto. South Asian J Res Microbiol. 2019;13:1-6.
- [CrossRef] [Google Scholar]
- Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264-74.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and molecular characterization of Staphylococcus aureus isolated from goats in Chongqing, China. BMC Vet Res. 2017;13:352.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular Characterization and antimicrobial susceptibility of Staphylococcus aureus isolates from clinical infection and asymptomatic carriers in Southwest Nigeria. PLoS One. 2015;10:e0137531.
- [CrossRef] [PubMed] [Google Scholar]
- SCC mec in staphylococci: Genes on the move. FEMS Immunol Med Microbiol. 2006;46:8-20.
- [CrossRef] [PubMed] [Google Scholar]
- Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J Clin Microbiol. 2009;47:2975-80.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcal cassette chromosome mec (SCCMEC) typing of methicillin-resistant Staphylococci obtained from clinical samples in south-south, Nigeria. World J Pharm Pharm Sci. 2016;5:91-103.
- [Google Scholar]
- Emergence of SCCmec Type I obtained from clinical samples in Shiraz Teaching Hospitals, South-West of Iran. Jundishapur J Microbiol. 2015;8:e16998.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterisation of methicillin-resistant Staphylococcus aureus (MRSA) isolated at a large referral hospital in Zambia. Pan Afr Med J. 2017;26:108.
- [Google Scholar]
- Prevalence and characterization of methicillin-resistant Staphylococcus aureus from community-and hospital-associated infections: A tertiary care center study. Antibiotics (Basel). 2021;10:197.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of SCCmec, SPA types and multi drug resistant of methicillin-resistant Staphylococcus aureus isolates among inpatients and outpatients in a referral hospital in Shiraz, Iran. BMC Res Notes. 2019;12:614.
- [CrossRef] [PubMed] [Google Scholar]
- Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323-36.
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic resistance and virulence gene characteristics of methicillin-resistant Staphylococcus aureus (MRSA) isolated from healthy Edible Marine Fish. Int J Microbiol. 2020;2020:1-9.
- [CrossRef] [PubMed] [Google Scholar]








